We are proud the announce that UPMEM, alongside leading R&D labs, is part of a successfully funded EU research project called BioPIM . With this EUR 3M grant, the connection between Processing In Memory and genomics should be durably established. Progress in both hardware and software are planned to target specific bioinformatics workloads and therefore tackle one of the most important computing sector of the coming decades.
The goal of BioPIM is the realization of cheap, ultra fast and ultra low energy mobile genomics that eliminates the current
dependence of sequence analysis on large and power hungry computing clusters/data centers commonly based on FGPA or GPUs.
BioPIM ambitions ultra-fast (>100x faster than currently available), energy-efficient (consuming >70% less energy than current platforms), and cost-efficient (>100x cheaper than servers) genome analysis a reality. This is about to transform microbial sequencing, genome sequencing, and exome sequencing in clinical settings and in the field so to guide diagnosis and treatment in mobile genomics.
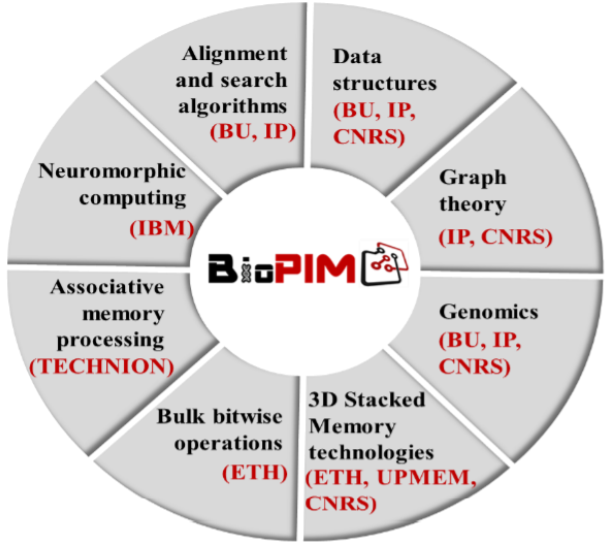
The targeted breakthrough of BioPIM is to push forward PIM architectures such as the UPMEM PIM to fundamentally improve the performance and energy efficiency of various important bioinformatics algorithms to make mobile genomics a reality, focusing on:
- in-memory alignment and search
- in-memory global and local genome assembly
- in-memory pangenome analysis
- in-memory variation calling and base calling.
The list of Partners in the BioPIM consortium:
- Bilkent(TR)
- ETH(CH)
- Institut Pasteur(FR)
- CNRS(FR)
- IBM(CH)
- TECHNION(IL)
- UPMEM(FR)
Key role and contributions